
Aashiq H. Kachroo, PhD.
- Associate Professor, Center for Applied Synthetic Biology, Biology
- Canada Research Chair Tier 2 in Systems & Synthetic Biology, Biology
- Member, Center for Structural & Functional Genomics, Concordia University., Biology
Are you the profile owner?
Sign in to editResearch areas: Systems Genetics & Synthetic Biology
Contact information
Email:
Availability:
Lab Phone 514-848-2424 ext. 5977
Website:
Biography
Education
PhD (Indian Institute of Science, Bangalore, INDIA)
PDF (The University of Texas at Austin, Austin, TX, USA)
Teaching activities
BIOL367-Fall
Molecular Biology
BIOL485/523/632-Winter
Agriculture & Agri-Food Biotechnology/ Advanced Biotechnology
Research activities
Engineering complex biological processes in simplified cells
The most deeply evolutionarily conserved human genes encode essential cellular machinery whose failures are linked to diverse diseases, from cancer to cardiovascular disease. Recent systematic studies have discovered extensive genetic polymorphism in these genes yet studying how these variations contribute to cellular function and overall human health remains a challenge. Our group is interested in studying these conserved human genes along with many of their variants, by systematically humanizing yeast cells, replacing each of the essential yeast genes in turn by its human version. The resulting strains will serve as new physical reagents for studying the human genes in a simplified organismal context, opening up simple high-throughput assays of human gene function, analyzing the impact of human genetic variation on gene function, the screening and repurposing of drugs and the rapid determination of mechanisms of drug resistance. Our work will illuminate fundamental principles of evolution, providing direct measurements of whether or not human and yeast genes have retained their ancestral functions over a billion years of evolutionary divergence.
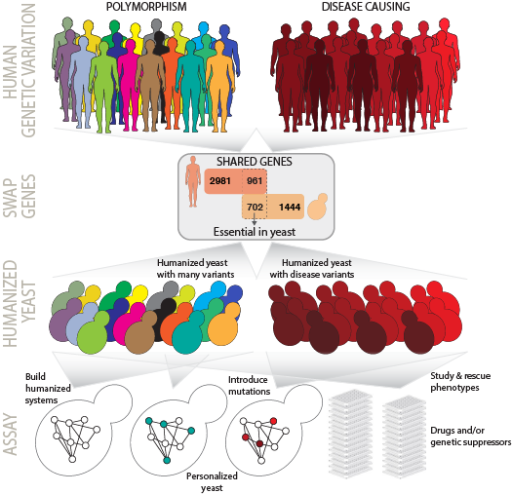
Humanized yeast - a platform for decoding human genetic variation and drug discovery
Aashiq H. Kachroo
Publications
- SELECT PUBLICATIONS
- * Equal author
- $Corresponding author
- Sultana S*, Mudabir A*, Li J., Hochstrasser M., Kachroo AH$. Species-specific protein-protein interactions govern the humanization of the 20S proteasome core in yeast. 2023 Genetics (Link) Preprint a bioRxiv (Link).
- Abdullah M&*, Greco BM&*, Laurent JM, Riddhiman KG, Boutz D, Vandeloo M*, Marcotte EM, Kachroo AH$. Rapid, scalable, combinatorial genome engineering by Marker-less Enrichment and Recombination of Genetically Engineered loci in yeast. 2023. Cell Reports Methods (Link), Preprint at bioRxiv (Link) In news (Link).
- Kachroo AH$, Michelle Vandeloo*, Greco BM*, Abdullah M*. Humanized Yeast to Model Human Biology, Disease and Evolution. Disease Models & Mechanisms. Epub 2022 Jun 6. (Link)
Participation activities
Undergraduate mentorship
Concordia University-iGEM supervisor
2018-present
2019- Gold Medal at iGEM 2019 & nomination for the Best Presentation
2020- Gold Medal at iGEM 2020 & Best Software Award
Research & Infrastructure funding
NSERC-Discovery; NSERC-CREATE; FRQNT; CFI; CRC

