RESEARCH
Design, synthesis, and physical characterization of bio-inspired model folding systems
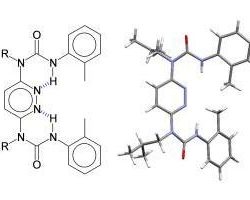
Organic synthesis is being used to access new macrocycles and helical foldamers that integrate the structural and functional characteristics of biopolymers with the stability and diversity of synthetic polymers. Scanning probe microscopies (STM and AFM) are being used to probe the self-assembly and mechanical properties of these novel materials.
Our research is directed towards the fundamental principles involved with molecular and macromolecular 'folding'. The objectives of this research program are the design, synthesis, and physical characterization of bio-inspired model folding systems (foldamers) that adopt well-defined shapes, conformations, and functions based on the sum of weak interactions.
Atomic force microscopy (AFM) has become a powerful tool for studying the mechanical unfolding and refolding of individual proteins and synthetic polymers. Polymeric helical foldamers are expected to have the property of being molecular-scale springs (nano-springs). We are currently using single molecule force spectroscopy to probe the physical properties of individual biopolymer molecules (chitosan) to determine their molecular-level adhesion and elastic properties.
In parallel we will perform steered molecular dynamics simulations to computationally stretch single foldamers and compare the resulting force-extension profiles with experimentally observed unfolding under external stretching forces. These experiments are yielding information into foldamer folding, unfolding, and refolding. Insight into protein folding and denaturation and in vivomechanical stretching, thought to play a role in regulating the function of proteins polysaccharides and DNA, will be gained. Applications of these 'molecular springs' for biotechnology and nanotechnology will be thoroughly explored.